ExoPlaSim Python API Documentation¶
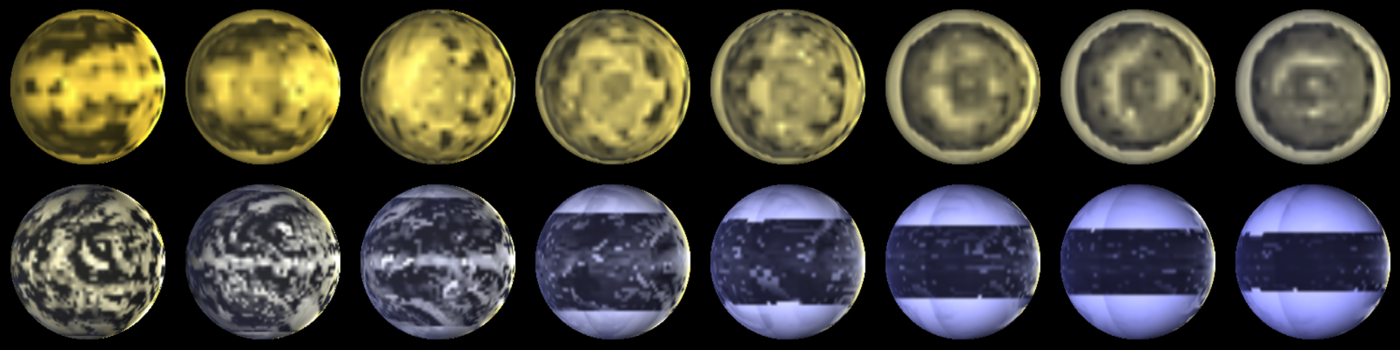
A range of planets modeled by ExoPlaSim, and postprocessed with SBDART. The top row consists of tidally-locked aquaplanets at T21 orbiting stars ranging from 2500 K to 4000 K, with orbital periods increasing with stellar mass. The bottom row consists of aquaplanets with 24-hour rotation at T42, orbiting stars ranging from 4000 K to 8000 K.¶
Contents¶
Created by Adiv Paradise
Copyright 2020, Distributed under the General Public License.
This API was written with Python 3 in mind, but should work with Python 2 and outdated versions of NumPy.
Requirements¶
Python (including development libraries, e.g. python-dev or python3.9-dev on Ubuntu–if using anaconda, these should already be included in your installation)
numpy
scipy (only needed for additional utilities, postprocessor)
matplotlib (only needed for additional utilities)
GNU C (gcc/g++) and Fortran (gfortran) compilers (development headers must be present)
(optionally) Other compilers whose use you prefer for the model itself
(optionally) MPI libraries for those compilers
Compatibility¶
Linux (tested on Ubuntu 18.04, CentOS 6.10): Yes
Google Colaboratory: Yes (note that OpenMPI support on Colaboratory is limited due to automatic root privileges; look up how to run OpenMPI executables with root permissions and note that this is not recommended)
Windows 10: Yes, via Windows Subsystem for Linux
Mac OS X: Yes, requires Xcode and developer tools, and OpenMPI support requires that Fortran-compatible libraries be built. Tested on Mac OS X Catalina and Big Sur (with MacPorts, GCC10, OpenMPI, and Anaconda3), Apple M1 compatibility has not been tested.
Optional Requirements¶
netCDF4 (for netCDF support)
h5py (for HDF5 support)
New in 3.2:¶
Experimental integration with petitRADTRANS to compute transit spectra and reflectance spectra, including maps and true-colour images (use at your own risk)
Ability to specify general keplerian orbits, with high eccentricity, using a revamped orbit code for higher accuracy
Orbital elements now included in standard output
Numerous bugfixes
New in 3.0:¶
ExoPlaSim no longer depends on X11 libraries for installation and compilation!
Revamped postprocessor no longer depends on NetCDF-C libraries, and supports additional output formats (including netCDF, HDF5, NumPy archives, and archives of CSV files).
GCC and gfortran support through GCC 10.
Improved cross-platform compatibility
Numerous bugfixes
Installation¶
pip install exoplasim
OR:
python setup.py install
If you know you will want to use NetCDF or HDF5 output formats, you can install their dependencies at install-time:
pip install exoplasim[HDF5]
OR:
pip install exoplasim[netCDF4]
OR:
pip install exoplasim[netCDF4,HDF5]
The first time you import the module and try to create a model
after either installing or updating, ExoPlaSim will run a
configuration script, write the install directory into its
source code, and compile the pyfft library.
You may also configure and compile the model manually if you wish
to not use the Python API, by entering the exoplasim/ directory
and running first configure.sh, then compile.sh (compilation flags
are shown by running ./compile.sh -h).
Most Common Error Modes¶
There are 3 major ways in which ExoPlaSim can crash. One is related to installation, one is related to model compilation/configuration, and one is related to numerical stability.
If in the run folder, diagnostic files are produced that appear to have made it all the way to the end of the year (there is a summary tag giving time elapsed and that sort of thing), then the problem is likely with the postprocessor. It is likely that the error output will be informative; if it is not clear how to resolve, please let me (the developer) know.
If the postprocessor itself is not the problem, then it’s likely you somehow passed incorrect output codes to the postprocessor. This is the most common scenario for postprocessor-related crashes. Check your inputs for any errors. In particular, note that climatology outputs are not available if storm climatology was not enabled.
If things crashed and burned immediately, it’s likely a configuration
problem. Check to make sure you aren’t using a restart file from a run
that used a different resolution, or stellar spectrum files that aren’t
formatted correctly (use the makestellarspec
utility to format Phoenix spectra for ExoPlaSim), or boundary condition
.sra files that aren’t properly-formatted.
If things were fine until they weren’t, then it’s likely ExoPlaSim encountered a numerical instability of some kind. Some of these are physical (e.g. you ran a model at a thousand times Earth’s insolation, and the oceans boiled, or the model was too cold and the physics broke), while some are not (something happened to violate the CFL condition for the given timestep, or an unphysical oscillation wasn’t damped properly by the dynamical core and it grew exponentially). If this happens, either try a model configuration that is more physically reasonable, or if the problem appears not to have been physical, try reducing the timestep or increasing hyperdiffusion. Sometimes it also works to slightly adjust a model parameter such as surface pressure by a fraction of a percent or less–just enough to nudge the model out of whatever chaotic local minimum it ran into, but not enough to qualitatively change the resulting climate.
New in ExoPlaSim 3.0.0, there is a “crash-tolerant” run mode. With this mode enabled, a runtime crash will result in rewinding 10 years and resuming. This deals with many of the most frustrating problems related to numerical instability. However, due to the potential for infinite loops, this is only recommended for advanced users.
PlaSim Documentation¶
Original PlaSim documentation is available in the exoplasim/docs/ folder.
Usage¶
To use the ExoPlaSim Python API, you must import the module, create a Model or one of its subclasses, call its configure method and/or modify method, and then run it.
An IPython notebook is included with ExoPlaSim; which demonstrates basic usage. It can be found in the ExoPlaSim installation directory, or downloaded directly here.
Basic example::
import exoplasim as exo
mymodel = exo.Model(workdir="mymodel_testrun",modelname="mymodel",resolution="T21",layers=10,ncpus=8)
mymodel.configure()
mymodel.exportcfg()
mymodel.run(years=100,crashifbroken=True)
mymodel.finalize("mymodel_output")
In this example, we initialize a model that will run in the directory
“mymodel_testrun”, and has the name “mymodel”, which will be used to
label output and error logs. The model has T21 resolution, or 32x64,
10 layers, and will run on 8 CPUs. By default, the compiler will use
8-byte precision. 4-byte may run slightly faster, but possibly at the
cost of reduced stability. If there are machine-specific optimization
flags you would like to use when compiling, you may specify them as a
string to the optimization argument, e.g. optimization='mavx'. ExoPlaSim
will check to see if an appropriate executable has already been created,
and if not (or if flags indicating special compiler behavior such as
debug=True or an optimization flag are set) it will compile one. We then
configure the model with all the default parameter choices, which means
we will get a model of Earth. We then export the model configurations
to a .cfg file (named automatically after the model), which will allow
the model configuration to be recreated exactly by other users. We
run the model for 100 years, with error-handling enabled. Finally, we
tell the model to clean up after itself. It will take the most recent
output files and rename them after the model name we chose, and delete
all the intermediate output and configuration files.
A Note on NetCDF and the (deprecated) Burn7 Postprocessor¶
As of ExoPlaSim 3.0.0, burn7 is deprecated. It is only
available via the exoplasim-legacy package.